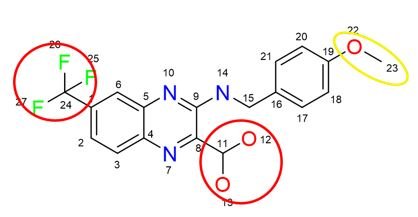
Molecular Complexity
What is Molecular Complexity?
1
The Quantitative Complexity Theory (QCT) bridges physics and information theory providing the first Dynamic Molecular Complexity (DMC) metric which quantifies the amount of information encoded in a particular structure. Analyzing the dynamics of atomic interactions in a molecule, the QCT pinpoints “complexity hotspots”/potential pharmacophores, which drive its biological function.
Molecular complexity is computed via a proprietary algorithm. By understanding the complexity of a molecule in the context of its dynamics, new knowledge and insights may be obtained. Conventional, “static” metrics are unable to quantify complexity correctly as it is not a constant. In fact, molecular complexity is impacted by external factors such temperature, pressure, or solvent (viscosity, pH), etc. This means that depending on the conditions, a molecule may exhibit a more or less complex behavior.
The bottom line: the complexity of a molecule cannot be inferred from its architecture alone. Complexity exists only in the context of dynamics, when a molecule vibrates and information is transmitted.
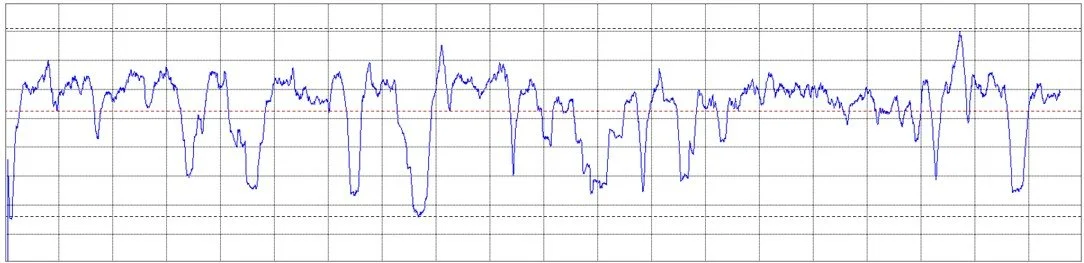
An example of a Molecular Complexity Map is illustrated in the image on the left. It reflects the multitude of interdependencies between atoms. The map changes over time and its topolgy fluctuates based on the actual molecular dynamics. An example of time history of molecular complexity is provided below.
Why is Molecular Dynamics so Important?
2
The biological and chemical properties of a molecule are fundamentally the expression of its dynamics. Molecules are dynamic, vibrating ensembles of atoms and their dynamics is not simply a background process; it is the very essence of information transmission and of biological function.
Our physics-based approach provides new insights into the intricate dynamics of molecules, identifying complexity hotspots, i.e., the regions and moieties that drive biological activity, indicating where to focus lead optimization. This can accelerate the pre-clinical trials phase by 50% to 75%.
The motion of atoms in a molecule is intricate and strongly nonlinear, as shown in the image on the left. Analyzing it via linear techniques, such as PCA, can potentially destort and destroy information. QCT, on the other hand, analyses molecular dynamics in its original, non-linear form, without assumptions or simplifications.
What makes BioDynLab different?
3
Current compound selection relies on synthesizability scoring and medicinal chemistry expertise rather than deterministic guidance for achieving optimization targets. This drives Chemical Space Exploration through trial-and-error. Without systematic prioritization, medicinal chemists must synthesize and test numerous analogs to identify successful modifications.
BioDynLab’s approach is deterministic and it distinguishes atoms essential for activity from those suitable for modification to improve ADMET properties without compromising potency. This is accomplished without the use of Machine Learning, meaning it is fast and bias free. Moreover, it offers 100% explainability.
BioDynLab’s QCT-powered OPTIMUS™ is used to analyse small molecules.
Can the QCT be used to Analyze Proteins?
4
Analyses of various proteins and peptides configurations through molecular dynamics simulations identifies critical amino acid hotspots that drive receptor binding and biological activity and can indicate best docking target sites. QCT analysis identifies also the key as well as the least significant drivers of the stability of its structure, opening the path to rational modification and design.
Preliminary research reveals that high-complexity amino acids are hydrophobic in nature, playing a key role in establishing the bonds that stabilize protein folding.
The images on the left show four mutations of the Alpha-1 antitrypsin protein, which have been analyzed via the QCT.
BioDynLab’s QCT-powered MACRO™ is used to analyse macromolecules.